Adhikary Lab
Gene Regulation
The central dogma of molecular biology is the flow of information from DNA to RNA (transcription), and from RNA to protein (translation). These processes are highly regulated at their discrete steps. Deregulated transcription is causal for a large fraction of human diseases including cancer and autoimmune disorders.
Protein binding to individual loci as well as transcript levels can be measured with high fidelity. Coupled to deep sequencing methods, expression and chromatin immunoprecipitation assays yield genome-wide datasets which give insight to regulatory principles. To characterise regulatory events in primary cells from human donors and in model cell lines, we furthermore use assays of upstream signaling events and signal integration at model gene loci, genetic approaches, and pharmacological intervention as well as proteomic high-throughput techniques. In vitro assays to measure modulation of enzymatic activity complement these approaches.PROJECTS
- Suppression of pro-inflammatory activation in the tumor microenvironment
- Negative gene regulation by nuclear receptors
Suppression of pro-inflammatory immune cell activation in the ovarian carcinoma microenvironment
Back to top
Macrophages are cells of the innate immune system which control tissue homeostasis, inflammation, and wound healing. The abundance of tumour-associated macrophages (TAMs) is correlated with poor prognosis in several solid tumour types. Ovarian carcinoma (OC) is frequently accompanied by a malignant peritoneal effusion termed ascites which harbours large numbers of floating cells, promotes metastasis, and, similar to plasma, has immunosuppressive properties conferred by soluble mediators. Like their counterparts from other tumour entities, TAMs isolated from OC ascites display a mostly immunosuppressive phenotype characterised by expression of markers such as the hemoglobin scavenger receptor CD163, TGFbeta1, and IL-10.
We found that the transcriptomes of ovarian carcinoma TAMs and those of peritoneal macrophages from non-tumour patients are very similar except fort a set of genes involved in extracellular matrix (ECM) reorganisation, a hallmark of wound healing and of tumourigenesis. TAMs can promote and perpetuate the resolution phase---the controlled decay of inflammation---which is in line with Virchow's notion that tumours are wounds that do not heal. Elevated expression of the ECM reorganisation gene set is negatively correlated with survival (Finkernagel et al. 2016), whereas increased expression of an interferon-inducible gene set in OC TAMs is positively correlated (Adhikary et al. 2017). Taken together, ovarian carcinoma TAMs are highly similar to peritoneal macrophages; discernible differences are limited to their numbers and expression of genes involved in ECM remodeling (pro-resolution) or IFN signaling (pro-inflammatory).
Immune evasion is an emerging hallmark of malignancy. A multitude of novel therapy approaches therefore relies on elicitation of tumouricidal action by immunocompetent stromal cells. Macrophage tumouricidal activity is exerted via secretion of cytokines, especially IL-12, which leads to T and NK cytotoxic functions. IL-12 stimulates production of interferon-gamma, a major determinant of the pro-inflammatory, anti-tumourigenic state. It is therefore desirable to enable and instigate IL-12 production in stromal macrophages. Tumors frequently exploit homeostatic mechanisms that suppress expression of IL-12. The p40 subunit of the IL-12 heterodimer, encoded by IL12B, is limiting for these functions. IL12B expression depends on interferon-gamma and the NFkB pathway.
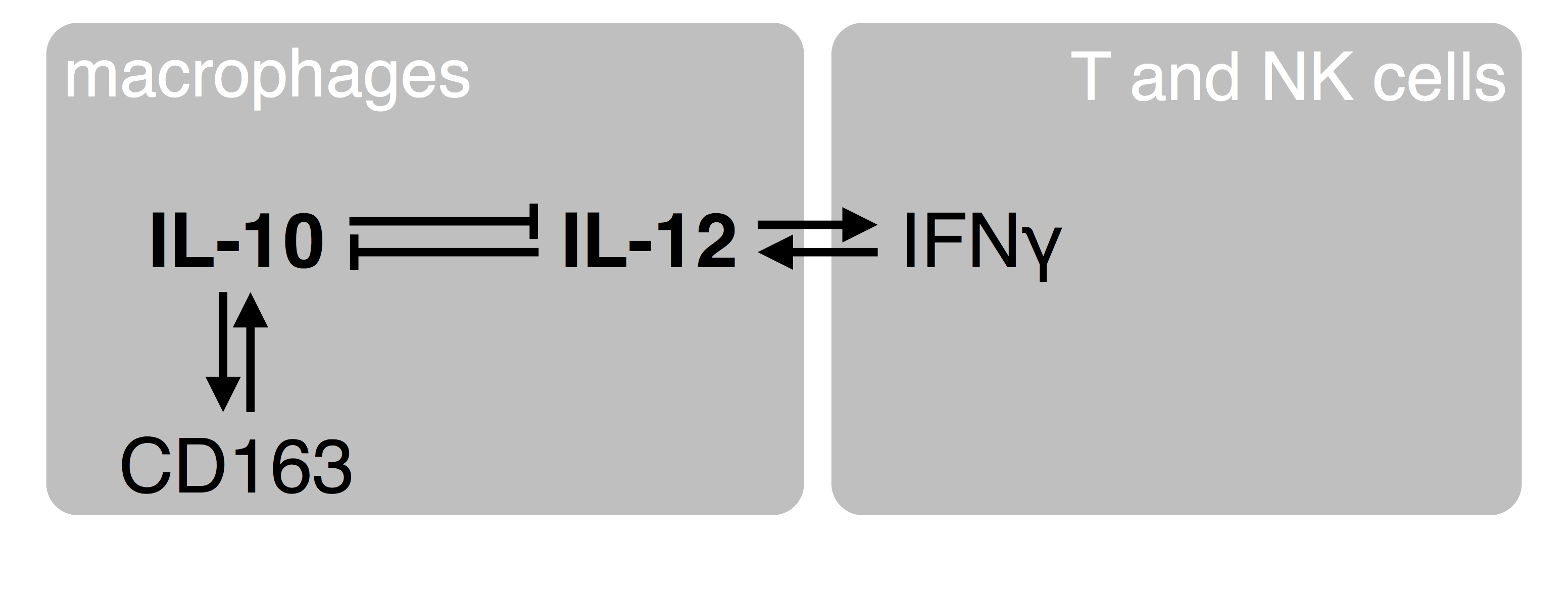
Figure 1. A simplistic view of crosstalk between macrophages and adaptive immune cells. The homeostatic IL-10 cytokine and the pro-inflammatory IL-12 reciprocally counteract their expression at the level of transcription. IL-10 enforces its own expression via the CD163 hemoglobin scavenger receptor and other signaling components. IL-12 enables the production of gamma-interferon by T and NK cells, which provides for positive feedback to the macrophages to increase synthesis of IL-12. Together, this serves as a flip-flop mechanism to switch between IL-10 (homeostasis; resolution of inflammation; wound healing; tumor immune evasion) and IL-12 (acute inflammation; tumor surveillance).
IL-10 alone is not sufficient to suppress expression of the IL12B gene which is limiting for production of IL-12 that can activate anti-tumorigenic cytotoxic functions of T and NK cells. We identified soluble factors which are necessary and sufficient to suppress transcription of IL12B by screening candidates derived from transcriptomic, proteomic, and lipidomic analyses. Surprisingly, these factors as well as ascites do not limit NFkB binding to chromatin, and other NFkB target genes remain readily inducible (Unger et al. 2018). In contrast to prevailing views, this indicates that the NFkB response is not generally defective in TAMs but rather modulated by gene-specific repression. We try to establish approaches that modulate IL12B expression, which could be of therapeutic value.
This project is carried out in collaboration with the groups of Rolf Müller and Silke Reinartz as part of the OvaRA consortium. Back to top
Negative gene regulation by nuclear receptors
Back to top
The nuclear receptor superfamily is a group of evolutionary related DNA-binding soluble proteins. Nuclear receptors (NRs) are recruited to direct target genes in a DNA sequence-specific manner mediated by zinc finger motifs of the DNA binding domain (DBD) of the receptor. NRs can bind small molecule ligands such as steroids and other lipids. These ligands enter the pocket structure of the ligand binding domain (LBD) and elicit conformational changes. Subsequently, different coregulator proteins are recruited to the receptor. In the nucleus, DNA-bound NR complexes regulate target gene expression in a ligand-dependent fashion, making them useful targets for pharmacological regulation of distinct gene sets. About 13% of clinically approved drugs target one or more of the 48 nuclear receptors encoded by the human genome.
Many natural and synthetic NR ligands are agonists. Agonists upregulate expression of direct target genes by inducing coactivator recruitment. Agonists can also transrepress the activation of pro-inflammatory gene expression via mechanisms that are incompletely understood. Inverse agonists are negative ligands which, in contrast to agonists, actively downregulate transcription of a subset of direct target genes of the respective receptor.
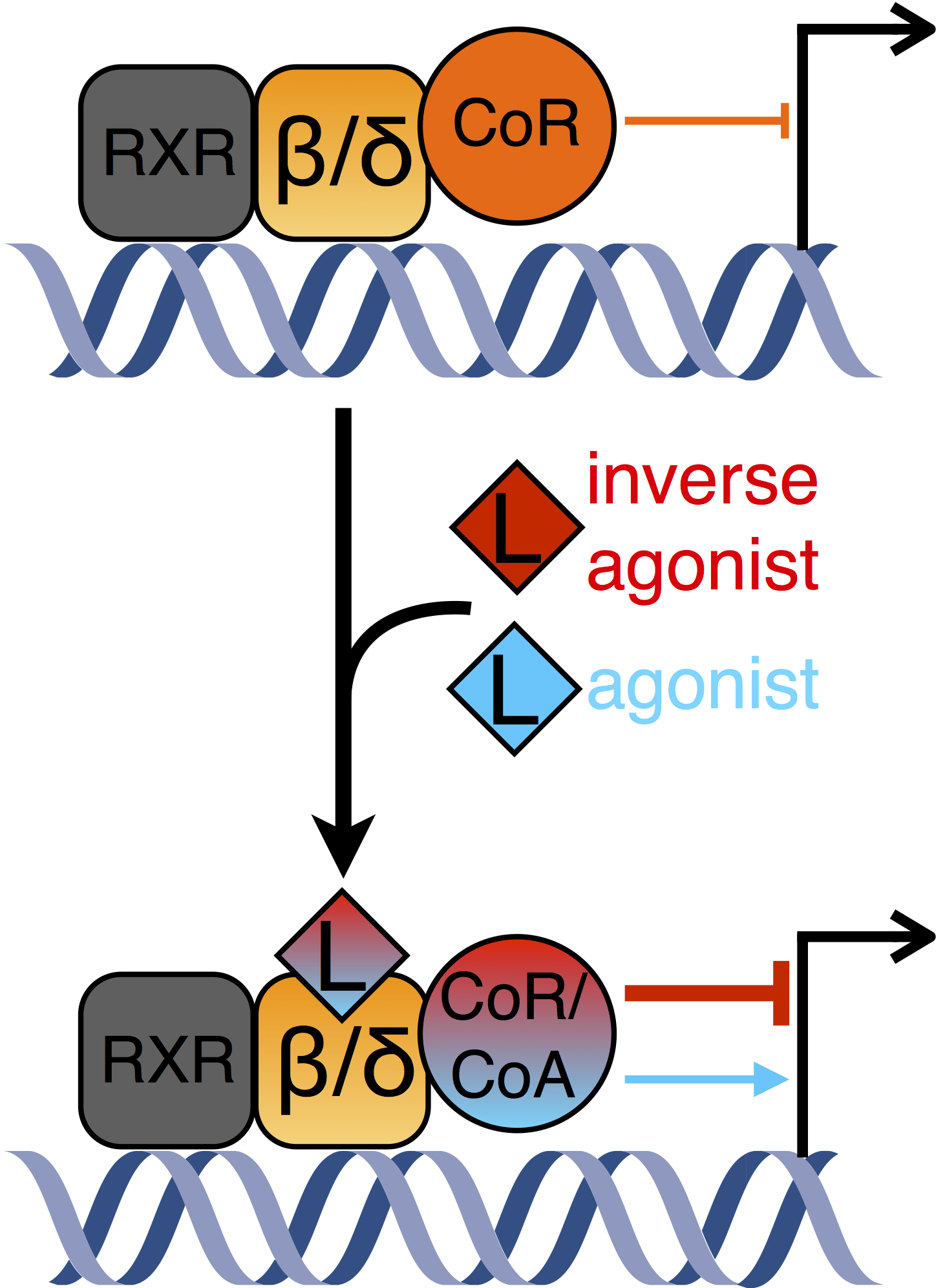

Figure 1. Left panel: A model of gene regulation by DNA-bound PPARβ/δ. In the unliganded state, corepressor proteins downregulate target gene expression. Basal repression can be alleviated by agonistic ligands. Inverse agonists lead to the recruitment of different corepressor complexes resulting in more pronounced repression of transcription. Right panel: Scanning chromatin immunoprecipitation analysis of the ANGPTL4 coding region. Cells were treated with different stimuli as indicated. The relative enrichment of DNA fragments obtained from a precipitation using an antibody against the large subunit of RNA polymerase II is plotted (see also Adhikary et al. 2013).
PPARβ/δ (peroxisome proliferator activated receptor beta/delta) is a ubiquitously expressed nuclear receptor which constitutively binds to DNA and regulates the expression of genes involved in processes such as wound healing and inflammation, fatty acid metabolism, and tumorigenesis. No clinically approved synthetic ligands are available for therapeutical use.
PPARβ/δ inverse agonists, which were developed by the Diederich group, are able to specifically downregulate expression of a subset of PPARβ/δ target genes. They abrogate induction of the model gene ANGPTL4 (angiopoietin-like 4) by different stimuli. Elevated ANGPTL4 expression is relevant in several pathophysiologies. Surprisingly, lysine deacetylase activity is not sufficient for repression, suggesting an uncharacterised mechanism (Adhikary et al. 2013).
NCOR and SMRT are corepressors that exist in multisubunit protein complexes harbouring HDAC3, which is a lysine deacetylase that has been suggested to be instrumental for repression by NCOR and SMRT.
We showed that recruitment of NCOR and SMRT complexes to PPARβ/δ is enhanced by inverse agonists. These complexes apparently prevent transcription initiation by interfering with binding of Mediator, a subset of general transcription factors, and RNA polymerase II. This is only partially dependent on lysine deacetylase activity (Legrand et al. 2019). Our findings indicate that NCOR and SMRT complexes use deacetylase-independent functions in addition to the activity of their HDAC3 subunit to downregulate transcription. These data shed light on mechanisms involved in transcription initiation and have strong implications on pharmacological intervention targeting NCOR/SMRT complexes.
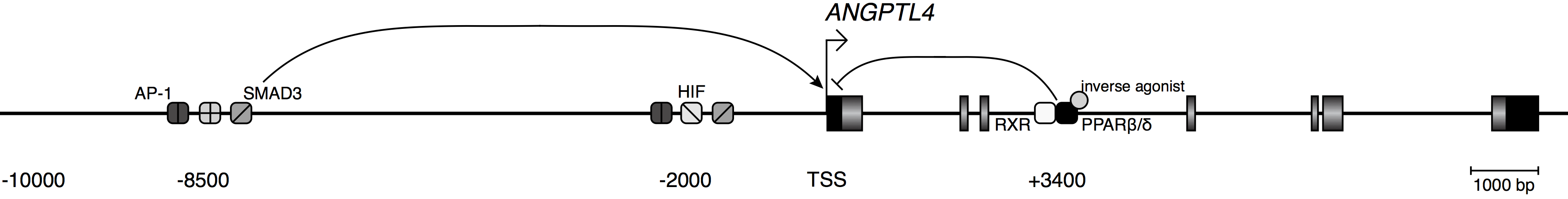
Figure 2. A model of the ANGPTL4 locus. Exons are depicted by rectangles. Black colouration marks the untranslated regions. TSS, transcription start site. Three enhancers, bound by the indicated transcription factors, integrate signals which lead to changes in expression level. PPARβ/δ inverse agonists lead to dominant repression of activating stimuli via limiting RNA polymerase II recruitment.
The compounds which specifically downregulate PPARβ/δ target gene expression (inverse agonists) are developed in close collaboration with the Wibke Diederich lab and synthesised in-house.
Back to top
Funding: Deutsche Forschungsgemeinschaft; Universitätsklinikum Marburg und Gießen GmbH
Last modified 28.02.2020

