Prof. Dr. Guntram Suske
The Sp transcription factor family
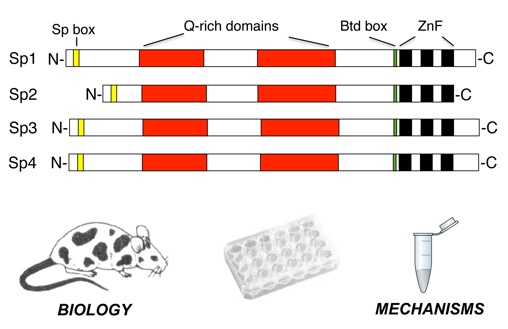
For long it is known that the zinc finger transcription factor Sp1 ("Specificity protein 1",
originally named according to a biochemical purification scheme that included Sephacryl
and phosphocellulose columns) binds and activates transcription through GC-rich promoter elements.
However, Sp1 is not the only transcription factor binding and acting through these elements.
It simply represents the first identified and cloned protein of a closely related protein
family characterized by a conserved zinc finger DNA binding domain. Mammals have nine
different Sp factors (Sp1 to Sp9), which can be grouped into two subclasses (Sp1 – Sp4
and Sp5 – Sp9) based on structural features outside of the zinc finger domain. Members of
the Sp1/Sp2/Sp3/Sp4 subclass share glutamine-rich activation domains. Sp1, Sp2 and Sp3 are
ubiquitously expressed raising important questions that are central to our research.
- What are the functions of these proteins in vivo?
- What determines their specificity?
- How are they regulated?
Projects
Sp transcription factors in development
We have investigated the biological functions of Sp2, Sp3 and Sp4 in the mouse by gene
targeting. Similar to Sp1null embryos [1], Sp2null embryos are severely growth-retarded
and die before embryonic day 10 [2]. Sp3null mice develop until birth but are not viable
due to manifold defects including impaired lung, cardiac, bone and red blood cell development
[3-6]. Compound heterozygosity of Sp1 and Sp3 results in embryonic lethality accompanied by a
spectrum of developmental abnormalities, including growth retardation, morphological alterations
of the lung, impaired ossification, anemia, and placental defects [7]. Sp4null mice are born
alive but two-thirds die within the first month after birth [8,9]. Sp4 is required for
specification of the cardiac conduction system [10,11] and normal brain development [12,13].
In conclusion, all four Sp factors have unique functions in vivo raising the question how
specificity is achieved.
SUMOylation of Sp3
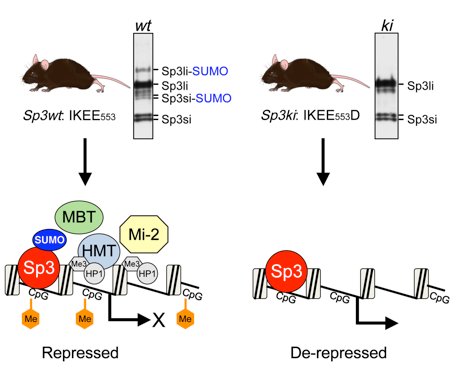
The activity of Sp3 is modulated by SUMOylation. SUMO modification of Sp3 occurs at a K551. SUMOylation inhibits the transcriptional capacity of Sp3 and represses transcription [14,15]. In a genome-wide RNA interference screen with cultured insect cells we have identified genes involved in SUMO conjugation and SUMO-mediated transcriptional repression [16]. Promoter-bound SUMO-modified Sp3 provokes the establishment of local repressive chromatin with features of compacted heterochromatin [17]. Finally, mice with a subtle mutation in the SUMO attachment site established SUMOylation of Sp3 as a molecular beacon for the assembly of repression machineries to maintain tissue-specific gene silencing [18].
Genomic binding of Sp1, Sp2 and Sp3
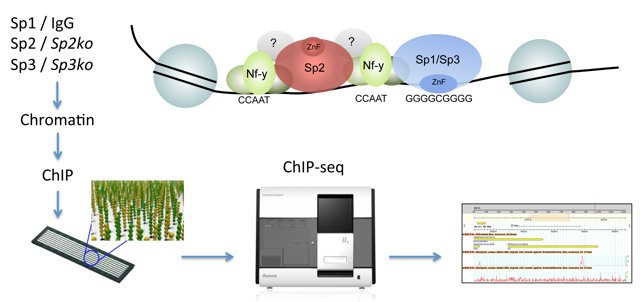
We have examined and compared the genomic binding patterns of Sp1, Sp2 and Sp3 in mouse embryonic fibroblasts and in a human cell line [19,20]. A key finding is that Sp2 does not localize at GC boxes as do the related transcription factors Sp1 and Sp3, but instead localizes at CCAAT motifs. Sp1 and Sp3 on the one hand and Sp2 on the other hand engage completely different protein domains for their genomic binding site selection. Most strikingly, the canonical DNA-binding domain of Sp2 is dispensable, and the N-terminal part sufficient for binding to its target promoters in vivo. We have identified the trimeric histone-fold CCAAT box binding transcription factor Nf-y as the major partner for Sp2-chromatin interaction. Nf-y is critical for recruitment of Sp2 to co-occupied regulatory elements. Equally, Sp2 potentiates binding of Nf-y to shared sites indicating the existence of an extensive Sp2-Nf-y interaction network. Altogether our results unveiled a striking and unexpected genomic loading mechanism of Sp2, and suggest that Sp2 acts as a cofactor rather than as a classical DNA-binding transcription factor.
References
1. Marin M, Karis A, Visser P, Grosveld F, Philipsen S (1997) Transcription factor Sp1 is essential for early development
but dispensable for cell growth and differentiation. Cell 89: 619-628.
2. Baur F, Nau K, Sadic D, Allweiss L, Elsasser HP, et al. (2010) Specificity protein 2 (Sp2) is essential for mouse
development and autonomous proliferation of mouse embryonic fibroblasts. PLoS One 5: e9587.
3. Bouwman P, Göllner H, Elsässer HP, Eckhoff G, Karis A, et al. (2000) Transcription factor Sp3 is essential for
post-natal survival and late tooth development. EMBO J 19: 655-661.
4. Göllner H, Dani C, Phillips B, Philipsen S, Suske G (2001) Impaired ossification in mice lacking the transcription
factor Sp3. Mech Dev 106: 77-83.
5. Van Loo PF, Bouwman P, Ling KW, Middendorp S, Suske G, et al. (2003) Impaired hematopoiesis in mice lacking the
transcription factor Sp3. Blood 102: 858-866.
6. Van Loo PF, Mahtab EA, Wisse LJ, Hou J, Grosveld F, et al. (2007) Transcription factor Sp3 knockout mice display
serious cardiac malformations. Mol Cell Biol 27: 8571-8582.
7. Krüger I, M. V, Simmons D, Elsässer H-P, Philipsen S, et al. (2007) Sp1/Sp3 compound heterozygous mice are not
viable: Impaired erythropoiesis and severe placental defects. Dev Dyn in press.
8. Supp DM, Witte DP, Branford WW, Smith EP, Potter SS (1996) Sp4, a member of the Sp1-family of zinc finger
transcription factors, is required for normal murine growth, viability, and male fertility. Dev Biol 176: 284-299.
9. Göllner H, Bouwman P, Mangold M, Karis A, Braun H, et al. (2001) Complex phenotype of mice homozygous for a
null mutation in the Sp4 transcription factor gene. Genes Cells 6: 689-697.
10. St Amand TR, Lu JT, Zamora M, Gu Y, Stricker J, et al. (2006) Distinct roles of HF-1b/Sp4 in ventricular
and neural crest cells lineages affect cardiac conduction system development. Dev Biol 291: 208-217.
11. Nguyen-Tran VT, Kubalak SW, Minamisawa S, Fiset C, Wollert KC, et al. (2000) A novel genetic pathway for
sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages.
Cell 102: 671-682.
12. Zhou X, Qyang Y, Kelsoe JR, Masliah E, Geyer MA (2007) Impaired postnatal development of hippocampal
dentate gyrus in Sp4 null mutant mice. Genes Brain Behav 6: 269-276.
13. Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, et al. (2005) Reduced expression of the Sp4 gene in
mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol
Psychiatry 10: 393-406.
14. Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, et al. (2002) Transcription factor Sp3 is
silenced through SUMO modification by PIAS1. EMBO J 21: 5206-5215.
15. Sapetschnig A, Koch F, Rischitor G, Mennenga T, Suske G (2004) Complexity of translationally controlled
transcription factor Sp3 isoform expression. J Biol Chem 279: 42095-42105.
16. Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, et al. (2008) Identification of SUMO-dependent
chromatin-associated transcriptional repression components by a genome-wide RNA interference screen.
Mol Cell 29: 742-754.
17. Stielow B, Sapetschnig A, Wink C, Krüger I, Suske G (2008) SUMO-modified Sp3 represses transcription
by provoking local heterochromatic gene silencing. EMBO Rep 9: 899-906.
18. Stielow B, Krüger I, Diezko R, Finkernagel F, Gillemans N, et al. (2010) Epigenetic silencing of
spermatocyte-specific and neuronal genes by SUMO modification of the transcription factor Sp3. PLOS Genet 6: e1001203.
19. Völkel S, Stielow B, Finkernagel F, Stiewe T, Nist A, et al. (2015) Zinc Finger Independent
Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It
from Sp1 and Sp3. PLOS Genet 11: e1005102.
20. Terrados G, Finkernagel F, Stielow B, Sadic D, Neubert J, et al. (2012) Genome-wide localization
and expression profiling establish Sp2 as a sequence-specific transcription factor regulating vitally
important genes. Nucleic Acids Res 40: 7844-7857.
